Hello everyone! I cannot believe that Friday was our last day in the lab! It has been such a great learning experience. In this time I have successfully extracted Dictyostelium genomic DNA and amplified the two flanking genomic regions surrounding the Dicty DDB_G0278957 gene, which encodes a putative mRNA decapping enzyme. I then cloned the flanking regions into a bacterial plasmid and introduced the plasmid to bacteria.
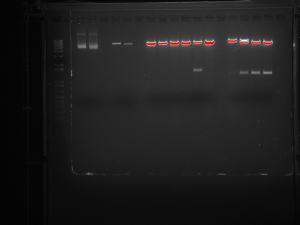
This picture shows the gel resulting from our restriction digest. This digest demonstrates that the bacterial plasmid DNA contains our insert. It was very exciting to see that all our work paid off.
We then used this plasmid to knock-out the Dictyostelium DDB_G0278957 gene by homologous recombination. The knock-out plasmid was engineered to confer blasticidin resistance and we were able to obtain D. discoideum colonies that were blasticidin resistant, suggesting that we were successfulin knocking out the DDB_G0278957 gene!. The next step is to obtain clonal isolates of the knock-out strain by diluting the cells and growing them on bacteria. The entire process of a Dicty transformation can take over a month! Once we confirm that the DDB_G0278957 is knocked out, we will analyze the biochemical consequences of the absence of the DDB_G0278957 gene and protein.
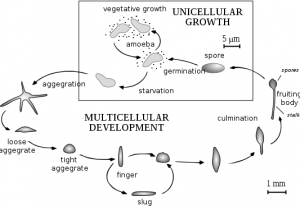
One of the coolest things we have done was to look at developing D. discoideum cells. When starved for nutrients, Dicty cells aggregate together to form a multicellular fruiting body containing spores that will be released when conditions are more favorable. The process of development takes 24 hours and we were able to induce development and see six developmental stages. We harvested these different stages to isolate mRNA and examine the expression pattern of DDB_G0278957 during development by Northern blotting. We started work early and plated the cells on non-nutritive filters. Throughout the day (and night), we looked at the plates under a dissecting microscope and observed the different developmental stages.. The first stage we observed was ripples. The cells gave the filters a shiny brown color and only four hours into development, waves could be seen forming on the plate. Four hours after that we saw loose mounds forming. By 11:30 at night these mounds had grown tips. It was really cool to see the cells changing. The next morning the cells had formed the final fruiting body stage containing the spores. Unfortunately, we did not have a camera set up to take pictures of the different stages as seen from a microscope but I have found this picture for you:
By Tijmen Stam, IIVQ (SVG conversion) – user:Hideshi (original version) (en:Image:Dicty Life Cycle H01.png) [GFDL (http://www.gnu.org/copyleft/fdl.html), CC-BY-SA-3.0 (http://creativecommons.org/licenses/by-sa/3.0/) or CC-BY-SA-2.5 (http://creativecommons.org/licenses/by-sa/2.5)], via Wikimedia Commons
Below you can see Dicty growing on plates containing bacteria. The clear regions are the regions where the Dicty cells have eaten the bacteria. As they deplete the bacteria, the cells aggregate and undergo development. The “fuzz” seen on the plates are the fruiting body containing spores.
This summer I have learned so much about molecular biology and have had a great time working with Kirsten and Dr. Parrish. Thank you Dr. Parrish and McDaniel for giving me such a great opportunity!