Math research can certainly be challenging, but when Dr. Hamblen told me that he, Sam, and I would be working with squares, I thought how hard could that be? That answer, very! We have been working away at using squared quaternions (basically one step more complicated than imaginary numbers) to make any quaternion, in as few steps as possible. As if this wasn’t enough, we are working with a value of i, which when squared is not a square. How can a square not be a square?? Luckily these questions are just as interesting as they are complicated, and this summer should be full of interesting discoveries. We are currently working with i squared is equal to negative two, and will continue to push our brains to think of new creative solutions!
Adventures in Hashawha
Ecological research is certainly full of excitement.
In our pursuit of knowledge, Meghan and I donned our waders, grabbed our nets, and traveled into the mysterious wilds of Lake Hashawha toward our target.
Our target you ask? The majestic dragonfly, or more specifically, their larvae.  These cricket-esque creatures are one of the top invertebrate predators in these types of aquatic environments, preying upon mosquito larvae, small tadpoles, and each other. Dragonfly larvae live mostly solitary lives, using substrate from their habitat to hide from predators and ambush prey.
These cricket-esque creatures are one of the top invertebrate predators in these types of aquatic environments, preying upon mosquito larvae, small tadpoles, and each other. Dragonfly larvae live mostly solitary lives, using substrate from their habitat to hide from predators and ambush prey.
Through our adventures in lake Hashawha, we encountered some of life’s most perplexing questions. Can one pull himself out of knee-deep mud pits using a friendly tree branch? Is crawling off of a dock into the mouth of an awaiting fish an evolutionarily advantageous behavior of dragonfly larvae? Do fish actually need water to survive?
Using our nets and a hint of cunning, we captured some larvae and placed them into our high-tech behavioral arenas to study activity level and hiding behavior.
With our arenas in the water and our behavioral trials commenced, we could finally begin the most annoying enjoyable aspect of our research; explaining to the countless families visiting Hashawha just what in the world we were doing.
Undergrad research – more fun for the professor?
Ever since I was a young undergraduate researcher at a small liberal arts college (SLAC), I knew that I wanted to work at a similar institution. Along the way, I have collaborated with some of the top scientists in my field, at large research-focused universities where a single researcher can get lost in a slew of graduate students and post-docs. Now that I’ve finished my first year teaching at McDaniel and I’m mentoring several undergraduate researchers, I know that I made the right choice.
Some of you might wonder “What’s so great about undergrad research?” Many PI’s (principal investigators; heads of research labs) prefer to have an army of researchers that are already trained to do the work on that lab’s specific project. Here are just some of the advantages of working with undergrads at McDaniel College.
Undergrad Enthusiasm
I am so impressed with the vigor with which my students dive into their projects. My style of mentoring is in line with the old adage “Give someone a fish, they eat for a day; teach someone to fish, they will eat for a lifetime.” I try to give my students the tools that they need to ask and answer their own questions. First I introduce them to a set of questions in line with my research program for which I think the student can yield some answers. I provide each student with some peer-reviewed literature to help them understand where their question “fits” into the broader picture and show them the techniques available to them in the Staab Lab.

Katelyn McCabe processing larval fish specimens; Paola Villegas examining slides that she stained yesterday; Rachel Statler optimizing high speed videos; Sophia Fricke and Nick Azat preparing solutions for a histological stain.
Once they have these resources, the sky is the limit! The research this summer has truly been student driven, while I am in the background providing support (materials, advice on methods, training in scientific communication, etc.). Indeed, many of the techniques that are occurring in the lab this summer were brought to me by curious students, rather than my forcing methods on them. More importantly, the students are thinking deeply about their projects, asking spin-off questions, and chasing after those answers. It is thrilling to mentor them through this process.
Resourcefulness and sense of community at a SLAC
When an experiment requires expensive equipment or materials, we question the purchase more carefully than someone at a Research 1 university. In fact, many techniques can be modified so that we can bypass expensive purchases This resourcefulness helps us gain a deeper understanding of the protocols and the functionality of the equipment.
For example, in the Staab lab, we process tissue for histological techniques. Tissue processors are machines that automatically transfer a piece of tissue from one solution to the next, without you having to think much about the process. These machines cost about $50,000. But we can manually transfer the tissue into each solution, and if we understand why we are using each solution, we can better monitor how the temperature, timing, and chemistry of the solution are affecting the tissue.
Not only that, but there is a sense of collegiality and sharing among the research labs.
Here is another more simple example of resourcefulness (and a request!): specimen jars can cost hundreds of dollars from scientific supply stores, but why not save those empty pickle/pasta sauce/olive jars for our pickled fishes? It’s true what they say about one person’s trash…
Honing my own scientific skills
Let’s be honest, some of that undergrad enthusiasm leads to big dreams of projects that could fill three Ph.D dissertations. It’s my job to reel in the students and help them realize how to completely answer one question before moving on to the next one. It is tempting for all of us to jump to the next shiny project (SQUIRREL!) but it is important to complete each part of the puzzle before building upon it.
The human element
My reasons for getting into science are likely much different than those at a large research university and as such, my responsibilities differ here at McDaniel. Rather than focusing on grant proposals and a high output of data, I am more concerned that my students obtain a rich learning experience that will prepare them for a lifetime of answering questions. Little might they know, they are providing me with a similarly rich learning experience.
Dicty and the DNA Drama
This summer, I don’t even miss my shorts and flip-flops. I prefer the lab coat, actually.
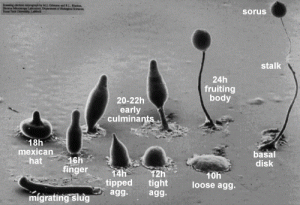
Dicty Fruiting Body Development
I have willingly traded summer essentials for lab gear and the opportunity to work with Dr. Parrish in her Dicty lab. We have spent the past three and a half weeks working on Dictyostelium discoideum. This social amoeba is fascinating, as it is unicellular when properly fed, but cells will aggregate and differentiate to form a multicellular fruiting body which will release spores once conditions improve. For my particular project, I am focused on a gene similar to the L534 gene in Mimivirus. This L534 gene is responsible for coding a mRNA decapping enzyme. My interest is in characterizing this similar gene in Dicty by observing the phenotype of Dicty without this gene.
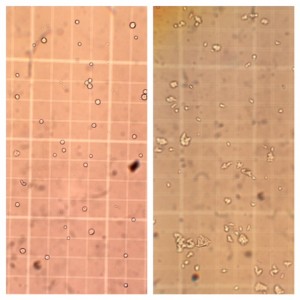
AX3** cell culture on left and L534 KO cell culture on right, on a haemocytometer for cell counting. (Click for a larger image.)
Previous research students have created the Dicty L534* knockout, and my task is to study the impact of the L534 gene’s absence to develop a better understanding of the gene’s function and importance in Dicty’s development and survival. There are three stages of my research which will provide details that we are seeking about the L534 gene in Dicty:
- Confirming the Knockout
- Observing Growth Rate
- Observing Cell Differentiation
The Dicty cultures have finally become healthy enough for completing these phases of the L534 knockout characterization, however, we have reached a snag in our master plan. To confirm the success of the knockout, we must extract genomic DNA from the Dicty and amplify it though PCR as well as visualize it through gel electrophoresis. We have completed the extraction and PCR, and we have run the gel, but much to our dismay, the DNA seem to have been degraded. Confirming the knockout is imperative to the continuation of the L534 characterization, as without the proper knockout of L534, we could not study the effects of the gene. And so, we have reached a determinant point. We will be repeating the genomic DNA extraction, PCR, and gel, and the outcome will either “make it or break it.” Stay tuned for the resolution to our obstacle…
*For convenience, we call the gene of interest in Dicty “L534,” though this is the name of the similar gene in Mimivirus rather than the actual name of the gene in Dicty. **The AX3 Dicty cells will serve as the wild type as a means of comparison. The AX3 cell culture is a strain of Dicty that can be grown axenically, meaning that it is grown on a nutritional substitute (HL5 medium) rather than the bacteria on which it normally feeds.
EPE Research: Florida and the First Training Week
During the last week in May I had the privilege of attending the annual conference of the American College of Sports Medicine (ACSM) in Orlando, Florida with Dr. McKenzie and Dr. Laird from our research team, along with my roommate for the trip, biomechanist, and avid Disney fan, Mr. Petrie. Dr. McKenzie and I presented a poster about our research from last summer. We hope everyone at the conference enjoyed their lesson about the most physiologically beneficial method for walking on an inclined treadmill. Some of the key points I learned are listed below:
- The ACSM annual meeting covers a broad range of topics and features a number of sessions on the basic sciences
- The conference itself is CRAZY with numerous sessions taking place at the same time and people dipping in and out of sessions at will.
- For better health, focus on being active, not on weight or waist circumference
- If you’re having trouble getting motivated to workout, make the activity a means to an end, rather the end itself (this includes taking stairs over elevators, standing more often, using your body as a mode of transportation- more than just your right foot, etc.)
During my week in Florida, the rest of the group faithfully finished all of the pre-testing such that we were able to stay on schedule and begin the training portion of our study this past week. For the training portion of the study, participants were matched and subsequently randomized into two different training groups operating under the following conditions:
- Standard training group- 3 min. at 60-65% maximum heart rate (max. HR), 27 min at 75-80% max. HR, 2 min. 60-65% max. HR
- High Intensity Interval Training (HIIT) group- 3 min. at 60-65% max HR followed by four bouts of alternating 4 min. at 90-95% max. HR and 3 min. at 60-65% max. HR
Both protocols are designed such that total work is the same. In both protocols, individuals arrive to the lab four times a week (Monday, Tuesday, Thursday, and Friday) to perform their training task for the day. The beauty of using heart rate to measure intensity is that it accounts for days when the participant is feeling “off” and it adjusts the intensity as fitness level increases. Already, we have noticed drastic differences in the ways that participants respond to the training load. Most of the participants in the standard training protocol appear to have adjusted fairly well; while only about half of the participants in the HIIT protocol appear to have adjusted, the other half are in visible distress throughout most of their training sessions. During one training session, a participant, having difficulty coping with the training, emphatically told me “I hate you Matt!”
Here’s to hoping we make it through another week of training visibly distressed HIIT participants.
-Matt Peterson (HIIT team)



