Here in the biology department, one of the best days of the summer research season is the summer research symposium – I love hearing about all the cool results from all the other labs, and also seeing my own students present their work to their peers. Here they are – the Biology summer 2014 crew!
Category Archives: Parrish Lab
Dicty and the DNA Drama
This summer, I don’t even miss my shorts and flip-flops. I prefer the lab coat, actually.
Dicty Fruiting Body Development
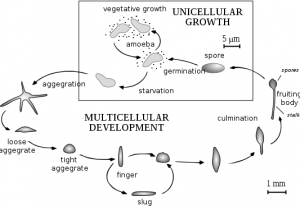
I have willingly traded summer essentials for lab gear and the opportunity to work with Dr. Parrish in her Dicty lab. We have spent the past three and a half weeks working on Dictyostelium discoideum. This social amoeba is fascinating, as it is unicellular when properly fed, but cells will aggregate and differentiate to form a multicellular fruiting body which will release spores once conditions improve. For my particular project, I am focused on a gene similar to the L534 gene in Mimivirus. This L534 gene is responsible for coding a mRNA decapping enzyme. My interest is in characterizing this similar gene in Dicty by observing the phenotype of Dicty without this gene.
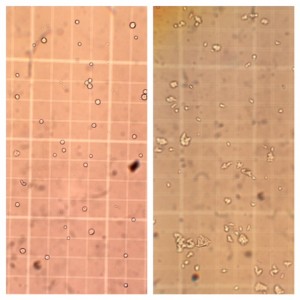
AX3** cell culture on left and L534 KO cell culture on right, on a haemocytometer for cell counting. (Click for a larger image.)
Previous research students have created the Dicty L534* knockout, and my task is to study the impact of the L534 gene’s absence to develop a better understanding of the gene’s function and importance in Dicty’s development and survival. There are three stages of my research which will provide details that we are seeking about the L534 gene in Dicty:
- Confirming the Knockout
- Observing Growth Rate
- Observing Cell Differentiation
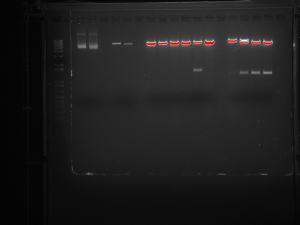
The Dicty cultures have finally become healthy enough for completing these phases of the L534 knockout characterization, however, we have reached a snag in our master plan. To confirm the success of the knockout, we must extract genomic DNA from the Dicty and amplify it though PCR as well as visualize it through gel electrophoresis. We have completed the extraction and PCR, and we have run the gel, but much to our dismay, the DNA seem to have been degraded. Confirming the knockout is imperative to the continuation of the L534 characterization, as without the proper knockout of L534, we could not study the effects of the gene. And so, we have reached a determinant point. We will be repeating the genomic DNA extraction, PCR, and gel, and the outcome will either “make it or break it.” Stay tuned for the resolution to our obstacle…
*For convenience, we call the gene of interest in Dicty “L534,” though this is the name of the similar gene in Mimivirus rather than the actual name of the gene in Dicty. **The AX3 Dicty cells will serve as the wild type as a means of comparison. The AX3 cell culture is a strain of Dicty that can be grown axenically, meaning that it is grown on a nutritional substitute (HL5 medium) rather than the bacteria on which it normally feeds.
Lord of the Dicty
Hello everyone! I cannot believe that Friday was our last day in the lab! It has been such a great learning experience. In this time I have successfully extracted Dictyostelium genomic DNA and amplified the two flanking genomic regions surrounding the Dicty DDB_G0278957 gene, which encodes a putative mRNA decapping enzyme. I then cloned the flanking regions into a bacterial plasmid and introduced the plasmid to bacteria.
This picture shows the gel resulting from our restriction digest. This digest demonstrates that the bacterial plasmid DNA contains our insert. It was very exciting to see that all our work paid off.
We then used this plasmid to knock-out the Dictyostelium DDB_G0278957 gene by homologous recombination. The knock-out plasmid was engineered to confer blasticidin resistance and we were able to obtain D. discoideum colonies that were blasticidin resistant, suggesting that we were successfulin knocking out the DDB_G0278957 gene!. The next step is to obtain clonal isolates of the knock-out strain by diluting the cells and growing them on bacteria. The entire process of a Dicty transformation can take over a month! Once we confirm that the DDB_G0278957 is knocked out, we will analyze the biochemical consequences of the absence of the DDB_G0278957 gene and protein.
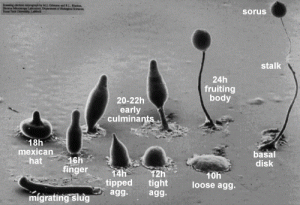
One of the coolest things we have done was to look at developing D. discoideum cells. When starved for nutrients, Dicty cells aggregate together to form a multicellular fruiting body containing spores that will be released when conditions are more favorable. The process of development takes 24 hours and we were able to induce development and see six developmental stages. We harvested these different stages to isolate mRNA and examine the expression pattern of DDB_G0278957 during development by Northern blotting. We started work early and plated the cells on non-nutritive filters. Throughout the day (and night), we looked at the plates under a dissecting microscope and observed the different developmental stages.. The first stage we observed was ripples. The cells gave the filters a shiny brown color and only four hours into development, waves could be seen forming on the plate. Four hours after that we saw loose mounds forming. By 11:30 at night these mounds had grown tips. It was really cool to see the cells changing. The next morning the cells had formed the final fruiting body stage containing the spores. Unfortunately, we did not have a camera set up to take pictures of the different stages as seen from a microscope but I have found this picture for you:
By Tijmen Stam, IIVQ (SVG conversion) – user:Hideshi (original version) (en:Image:Dicty Life Cycle H01.png) [GFDL (http://www.gnu.org/copyleft/fdl.html), CC-BY-SA-3.0 (http://creativecommons.org/licenses/by-sa/3.0/) or CC-BY-SA-2.5 (http://creativecommons.org/licenses/by-sa/2.5)], via Wikimedia Commons
Below you can see Dicty growing on plates containing bacteria. The clear regions are the regions where the Dicty cells have eaten the bacteria. As they deplete the bacteria, the cells aggregate and undergo development. The “fuzz” seen on the plates are the fruiting body containing spores.
This summer I have learned so much about molecular biology and have had a great time working with Kirsten and Dr. Parrish. Thank you Dr. Parrish and McDaniel for giving me such a great opportunity!
Come Dine with Dicty
Hi everyone!
Susan Parrish here. We are having a fun, albeit very busy summer here at the McDaniel Dicty lab. Kirsten and Catherine have really impressed me with their work ethic and they even have some exciting results! I will let them tell you about their results, since it is their work and I don’t want to steal their thunder. They are about to embark on project number two and the last few weeks will be very busy as they balance two projects at once.
One of my favorite aspects of being a college professor is inviting my research students over for dinner. I am fortunate to have a husband who is a wonderful cook, so this makes it much more enjoyable from my perspective. We had Catherine and Kirsten over for my husband’s fabulous BBQ last Thursday. He made his smoked BBQ brisket with grilled corn, asparagus, and squash. I made homemade baked beans, corn bread, and a cherry pie with local Baugher sour cherries. It was a veritable feast and a good time was had by all, including Bumble the Pug, our lab mascot.
Dicty Dancing
Hello everyone! My name is Kirsten Bickford and I am doing summer research with Catherine O’Keeffe and Dr. Parrish. We are using the amoeba Dictyostelium discoideum to eventually isolate putative mRNA decapping enzymes. These enzymes, termed Nudix enzymes, are found in large viruses all the way up to humans so being able to isolate and define the function of these enzymes is very exciting.
In order to do our experiments, we need a constant stock of healthy cells. We grow the Dicty cells on plates and in a flask and usually pass them about 3 times a week, so as to diminish genomic mutations. So far we have isolated genomic DNA, and are starting the process of doing a gene knockout. Currently, we are ligating and transforming parts of the gene into bacterial cells, but this is just the beginning of the process.
Compared to working in class laboratory settings, summer research is much more independent. Although I have learned sterile technique previously, there is much more pressure to do it properly as our cells will become contaminated otherwise. Many of the molecular techniques are new to me but with Dr. Parrish’s supervision, I have been able to learn how to do them properly and on my own. Pipetting is a huge factor and my technique has definitely improved since the start of the summer.
Hopefully our research will continue to go as planned, without equipment failures or major contamination. We will definitely be busy for the next 5 weeks, but another blog post is coming soon!